[ad_1]
Genetic variation and population substructure in Mexican Indigenous groups is influenced by geography
First, we compared our 716 Mexican Indigenous individuals from 60 ethnic groups (72 communities) with 146 previously published populations worldwide2,8,12, including Mexican Native American populations previously reported by Reich et al.8, Moreno-Estrada et al.2, and Silva-Zolezzi et al.12. The merged data set comprised 3490 individuals from 218 populations and 61,393 autosomal SNVs. Principal component analysis (PCA)13 indicated that Mexican Indigenous populations clustered with other Native American groups from North and South America (Supplementary Fig. 1). On the other hand, admixture14 analyses assuming K = 4 clusters showed that some Native American individuals are admixed with European and African populations, which is consistent with the history of the Mexican populations. We detected 325 Indigenous samples from the MAIS cohort with at least 0.99 Native American ancestry (Supplementary Fig. 2, upper panel).
In order to minimize the effects of recent admixture on our simulations, we performed local ancestry inference using RFMIX15 in each data set, except for Reich et al.8 data set as detailed in the “Methods” section. Non-Native ancestry tracks were masked in the individuals from Indigenous populations, and the masking accuracy was assessed by running the admixture analyses again assuming K = 4 clusters (Supplementary Fig. 2, lower panel).
Next, to assess the genetic structure of the Mexican Indigenous populations without the recent European and African ancestry, we combined the masked genomes of the Mexican Indigenous individuals from the MAIS cohort with the data sets from Reich et al.8, Moreno-Estrada et al.2, and Silva-Zolezzi et al.12, yielding a total of 1086 individuals. PCA in the whole Mexican Indigenous masked data set showed that the first axis of variation discriminated the Indigenous Mexican populations from the North, mainly groups from Aridoamerica, from those of Mesoamerica in the Center/South and Southeast (Fig. 1b). We also found a correlation between PC1 and the longitude and latitude (Supplementary Fig. 3a, b), and a Mantel test showed a significant correlation between genetic and geographical distances (p = 0.001, r = 0.63, Supplementary Fig. 3c). Moreover, PCA of Mesoamerican populations showed that the first two axes of variation separated the populations from the Center/South and Southeast following a geographic pattern (Fig. 1c). These results suggest that geographic location influences the genetic structure of these populations.
Furthermore, pairwise-FST comparisons identified the Tarahumara, Pima, Guarijio, and Cucapa in northern Mexico, and previously published populations, such as the Seri (North) and Lacandon (Southeast)2, had the highest levels of genetic differentiation when compared with the other populations based on this statistic (Fig. 2a and Supplementary Data 1). These observations suggest that these populations have experienced higher degrees of isolation or genetic drift, and possibly various founder effects that amplified this drift.
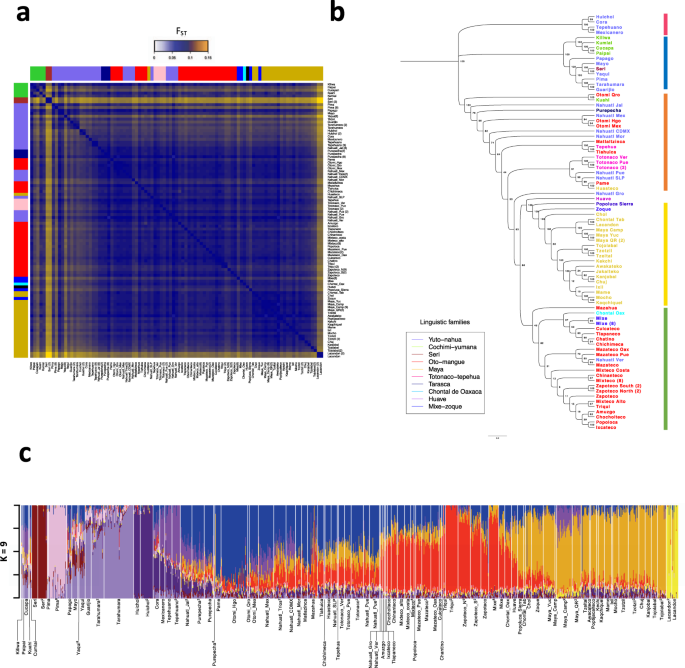
a Pairwise-FST matrix for all tested populations. Colored bars represent the linguistic family. b FST-based neighbor-joining tree showing the correlation with geographic location independent of linguistic classification (colored names), the numbers above the branches indicate the bootstrap values. Colored vertical lines represent the identified geographic regions: North (blue), Northwest (red), Central-east (orange), South (green), and Southeast (yellow). c Admixture analysis assuming K = 9 clusters in Mexican Indigenous populations. Superscript numbers are the corresponding references.
A midpoint rooted neighbor-joining (NJ) tree based on the pairwise-FST population distances showed a correlation between genetic structure and geographic distance, independently of the linguistic classification (Fig. 2b). The NJ tree topology revealed five major regions, with high clustering of the ethnic groups according to their geographic location. Furthermore, several ethnic groups from different regions are genetically closer to their geographical neighbors even if they belong to different linguistic families. For example, the Nahuatl from San Luis Potosi (Yuto-nahua), Pames (Oto-mangue), and Huasteco (Mayan) co-inhabiting the Huasteca region fall into the same clade from the NJ tree. Similarly, the Mixe (Mixe-zoque) inhabiting Oaxaca are closer to Oto-mangue linguistic family groups from Oaxaca and the Zoque (Mixe-zoque) from Chiapas are closer to Mayan linguistic family groups (Fig. 2a, b).
To better understand the genetic composition of Mexican Indigenous populations, we carried out a genetic clustering analysis with the unsupervised model algorithm ADMIXTURE14 using K = 2–16 clusters (Supplementary Fig. 4). The cross-validation procedure showed that, within the Mexican Indigenous populations, the K = 9 yields the lowest cross-validation error (Supplementary Fig. 5). Based on this K, six of these clusters were mainly observed in a single population (Seri, Tarahumara, Pima, Tepehuano, Huichol, and Lacandon). On the other hand, two of the clusters were mainly observed in several ethnic groups inhabiting the Center and South (here referred to as multi-ethnics), principally in populations from the Oto-mangue linguistic family, and the other cluster in populations from the Southeast that are part of the Mayan linguistic family. We observed that the multi-ethnic and Mayan components had opposite gradients, where the Mayan component was the most prevalent in the Southeast and the multi-ethnic components were more prevalent in the Center and South of Mexico (Fig. 2c and Supplementary Fig. 6).
Effective population size and divergence time estimation
To track the demographic histories of Indigenous Mexican populations, we estimated the effective population size (Ne) across time based on two different methods. We included 48 ethnic groups from the masked data set, all of them with sample sizes of at least 10 individuals (Supplementary Table 2). Demographic reconstructions based on linkage disequilibrium (LD) analysis16,17 showed little evidence of a fluctuation in Ne before 150 generations ago (Supplementary Fig. 7). To evaluate more recent demographic changes, we estimated the Ne based on identity by descent (IBD) tracks implemented in the IBDNe software18,19. We observed a decline in the Ne between 15 and 30 generations ago in all tested populations that overlaps with the beginning of the European colonization of the Americas, followed by an expansion (Fig. 3a and Supplementary Fig. 8).
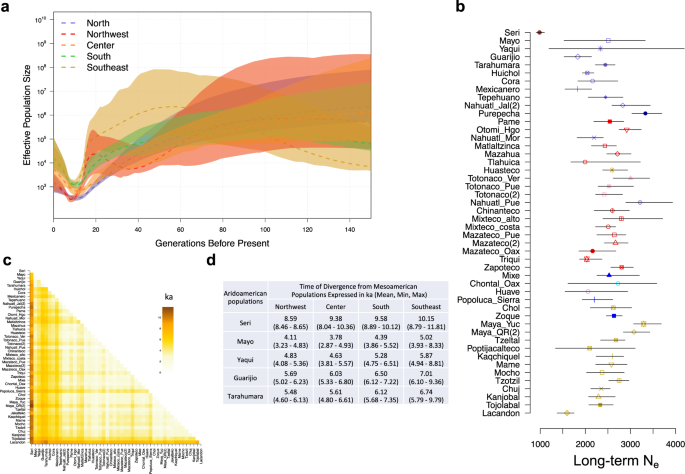
a Demographic measure of effective population size across time showing a decline in population sizes in the five main geographic regions identified here: North, Northwest, Center, South and Southeast. b Long-term Ne of all tested populations, shapes represent the long-term Ne and errors bars represent the 95% confidence interval. Colors are according to the legends in Fig. 1. Numbers between brackets are the corresponding references. c Divergence time between pairs of populations. d Mean of the observed T between Aridoamerican and Mesoamerican populations expressed in ka.
Next, we estimated the long-term Ne based on LD patterns using Neon Software16,17. The long-term Ne calculated in the whole sample set was 3169 (confidence interval of 2952–3402), which is similar to previous findings5,20,21. However, here we documented a variation in the long-term Ne between ethnic groups (Fig. 3b and Supplementary Table 3). The long-term Ne was smaller in highly differentiated populations, such as Seris and Lacandons (984 and 1593, respectively). Other ethnic groups had a long-term Ne between 1825 and 3331 individuals (Supplementary Table 3) and are similar to those previously reported in populations such like Tarahumara, Huichol, Triqui, and Maya22. The smaller long-term Ne may have contributed to greater genetic drift and lower genetic diversity in these ethnic groups. To confirm this hypothesis, we inferred autozygosity using runs of homozygosity (ROH). As expected, the Seri and Lacandon groups had the highest proportion of the genome in ROH compared to the other populations tested, suggesting that the high genetic differentiation observed in these populations is due to genetic drift as previously reported2 (Supplementary Fig. 9). We did not observe this phenomenon for other divergent populations, such as the Cucapa, Tarahumara, Guarijio, Tepehuano, and Huichol. In addition, the categorization of ROH by size showed that all tested Native American populations have a high proportion of short ROH (1–2 Mb), which is consistent with the fact that these populations have experienced a series of bottlenecks throughout their history23,24. Moreover, with the exception of Yaqui, Mazateco from Oaxaca, Chontal from Oaxaca, and Maya from Yucatan and Quintana Roo, we observed that all tested populations exhibited different proportions of ROH longer than 8 Mb (Supplementary Fig. 10), which is consistent with the presence of episodes of isolation and/or inbreeding23,24.
Both the long-term Ne and FST between pairs of populations were employed to calculate the divergence time between populations in generations (T) assuming a clean split between them. To scale T in years, we assumed 28 years per generation25. Seri and Lacandon populations have the highest T values compared to other populations (Fig. 3c and Supplementary Data 2), and the uppermost value of T was observed between Seri and Maya from Quintana Roo (T = 11.8 ka ago, Supplementary Data 2). Considering the ecogeographic region, we observed a higher T between populations from different regions than those from the same region. Populations from northern Mexico corresponding to Aridoamerica diverged from the populations in the Center/South around 3.96–9.47 ka ago and from the Southeast populations ~4.84 to 10.15 ka ago (Fig. 3c, d and Supplementary Data 2).
To better understand the demographic connections among the Mexican indigenous populations, we performed an IBD analysis in 325 individuals from our data set with >99% Native American ancestry using Hap-IBD26 (see “Methods” section) (Supplementary Table 4). We also explored the ethnic group genetic affinities within and between different geographical regions according to those observed in the NJ tree and defined previously by Contreras-Cubas et al.9. In line with that observed with allele frequency-based methods (Supplementary Fig. 3), the IBD analysis also showed that the indigenous populations are related to each other following an isolation by distance model, both at the intra and interregional level. Therefore, in most cases, neighboring indigenous populations are more likely to relate to each other than to distant groups (Fig. 4 and Supplementary Fig. 11). At the intraregional level, this trend is exemplified by Tarahumara and Guarijio from North (Fig. 4a) or Chuj and Kanjobal from Southeast (Fig. 4e). Additionally, the shared IBD segment analysis revealed gene flow between Indigenous populations from different regions in Mexico. Some examples with shared IBD blocks were observed between Cora from Northwest and Zapoteco from South or Guarijio, Tarahumara and Seri from the North and Mayan groups from the Southeast (Supplementary Data 3–7).
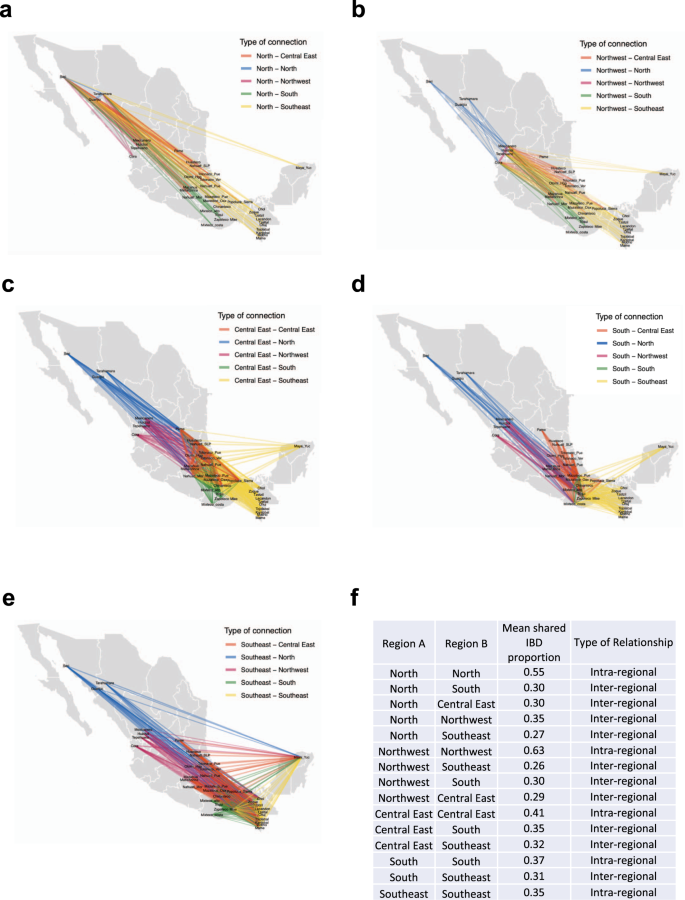
IBD segments analysis performed in Indigenous populations with at least 99% of Native American ancestry inferred from Admixture K = 4. Analyses were restricted for segments >7 cM. Shared IBD fragments shown proximal and distal connections between populations from the same and different regions. a North IBD segments, b Northeast IBD segments, c Central East IBD segments and d South IBD segments and e Southeast IBD segments. The width of each edge is proportional to the mean IBD length. f Table showing the mean values of IBD sharing within and between regions.
An IBD analysis incorporating all populations per region using both intermediate (5–10 cM) or large (>10 cM) shared IBD blocks revealed possible spatiotemporal interaction dynamic patterns among indigenous groups. Intermediate IBD block sizes are suggested to be dated to 500–1500 years ago (oldest), while large tracks are thought to be originated 0–500 years ago (youngest)27.
Analysis of intermediate block sizes revealed that the Central East, South, and Southeast regions have older connections among them than do the northern regions (Supplementary Fig. 11a). Meanwhile, large IBD track analysis suggested that the North region has a more recent gene flow with Northwestern and Central East regions than do South and Southeast regions (Supplementary Fig. 11b).
Genetic affinities between modern Native American populations and ancient inhabitants of the Americas
To gain more insight into the early migration patterns, we compared the previously published genomes of the Anzick-128 and Upward Sun River 1 (USR1)29 individuals with our data from the most representative sample of Indigenous peoples in the Mesoamerican and Aridoamerican regions of Mexico to date. First, we compared the ancient genomes with 59 worldwide populations and 325 individuals from our data set with at least 99% Native American ancestry (Supplementary Table 4) using an outgroup f3-statistic in the form of f3(Yoruba; Ancient, Modern). This analysis showed a high affinity of both ancient genomes with present-day Mexican Indigenous samples (Supplementary Fig. 12 and Supplementary Data 8).
We then combined the 325 indigenous samples with seven ancient genomes from American and South American populations3,30 (Supplementary Table 5), yielding a total of 111,586 autosomal SNVs. A TreeMix tree on this data set placed the USR1 genome at the basal position of all Native American populations tested, including Anzick-1. Meanwhile, all Aridoamerican populations formed a separate clade from those formed by the Mesoamerican populations and Anzick-1 and the NNA/ANC-B branch. Similarly, a PCA including the ancient samples showed that the Anzick-1 genome is more closely related to Mesoamerican populations than Aridoamerican populations, whereas USR1 is placed as an outlier in the PCA space (Supplementary Fig. 13).
The TreeMix tree analysis suggested a deep divergence between populations in Aridoamerica and Mesoamerica (Fig. 5a and Supplementary Fig. 14) prior to the divergence between Mesoamerica and the Anzick-1 individual. This observation is inconsistent with T < 10 ka, as calculated based on FST and long-term Ne (Fig. 3c), and the fact that a TreeMix tree allowing 20 migration edges (Supplementary Fig. 14) and the IBD networks analyses (Fig. 4) exhibited multiple gene flow between Mesoamerican and Aridoamerican populations. To test this, we calculated a D-statistic in the form of D(Yoruba, NNA/ANC-B; AA, MA) using the Ancient Southern Ontario population Canada_Lucier31 and Athabaskan32 ancient genomes as representatives of NNA/ANC-B ancestry. We found these results to be consistent with D ~ 0, suggesting that the NNA/ANC-B populations are an outgroup for those from Aridoamerica and Mesoamerica from Mexico (Supplementary Figs. 15 and 16), which is consistent with the TreeMix tree. To test whether Mesoamerican populations form a clade with Anzick-1 to the exclusion of Aridoamerican populations, we estimated a D-statistic in the form of D(Yoruba, AA; Anzick-1, MA). We found that Mesoamerican populations share more alleles with Aridoamerica than Anzick-1 shares with Aridoamerica (Supplementary Fig. 17).
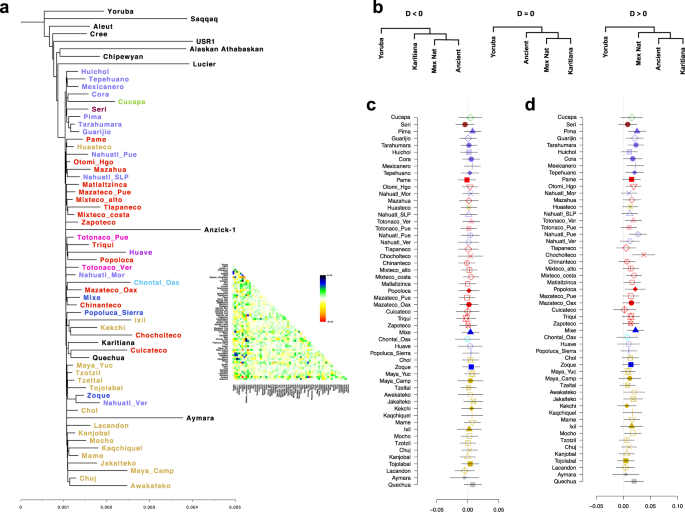
a Maximum-likelihood tree inferred from allele frequency, residual plot from the maximum-likelihood tree is shown. b Possible tree topologies resolved by D-statistics. c D-statistic in the form of D(Yoruba, USR1; Mex Nat, Karitiana) shows that all Mexican populations are related to the USR1 genome. d D-statistic in the form of D(Yoruba, Ancient; Mex Nat, Karitiana). Error bars in c, d represent 3 standard errors estimated by a weighted block jackknife. Mex Nat Mexican Native population.
To further explore the relationships between the Indigenous Mexican populations and the ancient samples, we estimated a D-statistic in the form of D(Yoruba, Ancient; Mexican Native population, South American), where the ancient samples were either USR1 or Anzick-1, and the South American samples were either Karitiana or Aymara. We also included the Tzotzil population as an internal control. When USR1 was used to represent the ancient population, we did not observe any significant deviation from zero in any of the tests (Fig. 5b and Supplementary Fig. 18a, b). However, when Anzick-1 was used in the test, we found that Anzick-1 shares more alleles with the South American population than with some of the Indigenous Mexican populations. This is particularly the case for populations from Aridoamerica, with the exception of Cucapa and Seri, as well as some Mesoamerican populations, such as Totonaco from Veracruz, Nahuatl from Puebla, Otomi from Hidalgo, Mixteco Costa, Chocholteco, Mocho, and as previously observed Mixe. Although we only found significant results (|Z| ≥ 3.2) when Karitiana and Tzotzil were used in the comparison, we observed a similar trend when Aymara was used in the test (Fig. 5c and Supplementary Fig. 18c, d). These results suggest that some of the Indigenous populations in our data set carry ancestry from a population that split before the Anzick-1 individual. Previous studies have suggested that the Mixe carry additional ancestry from an unknown population related to the SNA/ANC-A branch that split above the Anzik-1 individual (UPopA)33. Our results are consistent with this observation, suggesting that other populations from Aridoamerica and Mesoamerica may carry this ancestry (Fig. 5c and Supplementary Fig. 18c, d).
[ad_2]
Source link



/cloudfront-us-east-2.images.arcpublishing.com/reuters/QX63CELGCNKK5ND2AOWZ4WFSVQ.jpg)









